SUMO modification regulates the Drosophila innate immune response
Bhagyashree Kaduskar, Amarendranath Soory, Prajna Nayak, Sushmitha Hegde, Neel Wagh, Girish Ratnaparkhi
Post-translational modifications such as phosphorylation and ubiqutination have been demonstrated to play critical roles in attenuating signal transduction pathways and regulating expression of the defense genes [1,2,3,4]. SUMOylation is another such post-translational modification that has been shown to play a role in host defense. However, only a few proteins have been identified as bonafide SUMO targets. In the innate immune system, these bonafide targets include, the Drosophila NF-kappaB/Dorsal protein and products of Dif, IB and Nemo genes [5,6,7]. Studies done on amorphic and hypomorphic mutants of the SUMO conjugating enzyme Ubc9 [8] have shown that SUMOylation negatively regulates Toll/NFkappa-B signaling leading to a decrease in the transcriptional activation of the AMP gene Drosomycin (Drs). Ubc9 mutants also exhibit an over-proliferation of hemopoietic cells [8] suggesting a function in the cellular immune response. These studies along with the identification of some of the SUMOylated components of the Toll/ NFkappa-B and IMD pathways [5,6,7] indicate that SUMOylation is important in host defense.
Given the importance of a robust innate immune response and the widespread nature of this phenomenon, it is likely that many more proteins and signal transduction pathways involved in host defense are subject to regulation by SUMOylation. Identification of these proteins using a proteomics based screen and understanding their regulation by SUMO modification will help provide further insights into the mechanisms regulating host defense.
Our Goals are to conduct a detailed study on the effect of SUMOylation in the innate immune response by (i) Characterizing the effect of SUMO loss of function on animal host defense. (ii) Generating a parts-list of SUMOylated proteins that show change in their SUMO status on infection using a proteomics based approach and (iii) Decipher the diverse functional roles of SUMO in regulating host defense both at the level of the individual protein and also at the systems level.
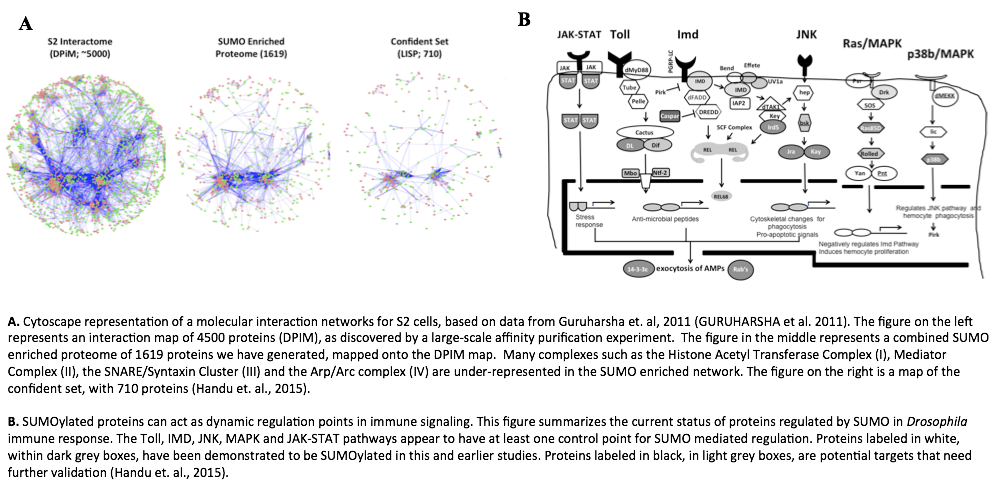
With the above goals in mind we have conducted a quantitative proteomics screen to list a set of 702 proteins whose SUMOylation state changes in S2 cells, on infection [8]. We [8] and others [9][10] have already shown that loss of SUMOylation can modulate the immune response in flies. Currently, we are using CRISPR/Cas9 based genomic editing to generate mutants of proteins that are SUMOylated in response to infection. We have successfully generated transgenic fly lines where target proteins are resistant to SUMOylation.
1. Ghosh, S. and D. Baltimore, Activation in vitro of NF-kappa B by phosphorylation of its inhibitor I kappa B. Nature, 1990. 344(6267): p. 678-82.
2. Chen, Z., J. Hagler, V.J. Palombella, F. Melandri, D. Scherer, D. Ballard, and T. Maniatis, Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev, 1995. 9(13): p. 1586-97.
3. Bhoj, V.G. and Z.J. Chen, Ubiquitylation in innate and adaptive immunity. Nature, 2009. 458(7237): p. 430-7.
4. Golebiowski, F., I. Matic, M.H. Tatham, C. Cole, Y. Yin, A. Nakamura, J. Cox, G.J. Barton, M. Mann, and R.T. Hay, System-wide changes to SUMO modifications in response to heat shock. Sci Signal, 2009. 2(72): p. ra24.
5. Nie, M., Y. Xie, J.A. Loo, and A.J. Courey, Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One, 2009. 4(6): p. e5905.
6. Bhaskar, V., M. Smith, and A.J. Courey, Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol Cell Biol, 2002. 22(2): p. 492-504.
7. Huang, T.T., S.M. Wuerzberger-Davis, Z.H. Wu, and S. Miyamoto, Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell, 2003. 115(5): p. 565-76.
8. Handu M, Kaduskar B, Ravindranathan R, Soory A, Giri R, Elango VB, Gowda H, Ratnaparkhi GS. SUMO-Enriched Proteome for Drosophila Innate Immune Response. G3 (Bethesda). 2015 Aug 18;5(10):2137-54. doi: 10.1534/g3.115.020958.
9. Chiu H, Ring BC, Sorrentino RP, Kalamarz M, Garza D, Govind S. dUbc9 negatively regulates the Toll-NF-kappa B pathways in larval hematopoiesis and drosomycin activation in Drosophila. Dev Biol. 2005 Dec 1;288(1):60-72. Epub 2005 Oct 24.
10. Paddibhatla I, Lee MJ, Kalamarz ME, Ferrarese R, Govind S. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 2010 Dec 23;6(12):e1001234. doi: 10.1371/journal.ppat.1001234.